May 2010 archive June 2010 archive
June 2010 archive July 2010 archive
July 2010 archive
07/30/10
Started to implement the new StaticDNASequence. The build process is going to get a little ugly, but the ranks should still be relatively clean (which is what matters). To make it easier on myself, I'm just going to do one pass through the whole sequence first to find all the N's and build the new data structure, then build the rest of the rank structure (rank blocks + count blocks). This is kind of wasteful because I can actually build the $ structure while building the rank structure (for just one pass), but I think it might be harder, given my current code. So, the build process will be a bit slower (which I think is okay...).
Finally, I did some benchmarks on the
count function. Test parameters are:
- read length = 35
- reference = e coli genome NC_010473, minus two characters (there was one Y and one R...) (length = 4,686,136 bases)
- number of reads = 10,000,000, generated randomly
- this randomly generated set was copied 10 times, for a total of 100,000,000 calls to
count.
- random seed = 1234
- times are reported in milliseconds wall time.
- Build version is 64-bit, Release mode (
-O3)
- Memory usage is just the memory used by the rank structure
| Sampling rate (bases) |
Memory usage (bytes) |
Type |
SSE 4.2? |
Time (ms) |
| 64 |
1,784,768 |
2-bit |
No |
27,212 |
| 256 |
1,339,168 |
2-bit |
No |
46,663 |
| 64 |
2,973,048 |
3-bit |
No |
31,454 |
| 256 |
2,081,848 |
3-bit |
No |
52,876 |
| 2048 |
1,821,912 |
3-bit |
No |
156,832 |
| 64 |
1,784,768 |
2-bit |
Yes |
19,924 |
| 256 |
1,339,168 |
2-bit |
Yes |
33,002 |
| 64 |
2,973,048 |
3-bit |
Yes |
24,371 |
| 256 |
2,081,848 |
3-bit |
Yes |
38,524 |
| 2048 |
1,821,912 |
3-bit |
Yes |
90,328 |
For a (very) rough comparison,
saligner, using readaligner's FM index, takes 64.461 seconds to do an exact mapping for 2.7 million reads (each 35 bases in length). This means it would take roughly 2,387 seconds to align 100,000,000 reads. However, this is not a very good comparison because that one's doing a
locate function which is more complex, has to read from a file and write to an output file, and runs at 32 bits. Also, the rank structure might use a different sampling rate, although I believe it uses a sampling rate of 256, which is why I included that test in. So, this means we have quite a bit of room to work with!
Another note is that for the 3-bit structure, it requires a sampling rate of 2048 to be roughly comparable in memory usage. However, at this rate, it's much slower than the 2-bit structure (roughly 5-6 times slower).
To do:
- Finish the $ data structure implementation in the StaticDNASequence
08/04/10
Spent all of yesterday and most of today finishing up the $ data structure for the StaticDNASequence class. I've finished testing it thoroughly (I hope). I've also integrated it with the FM Index and tested the count method to verify the output. Basically, I used the
std::string::find method to look through a reference sequence (I used
NC_010473, the E. coli genome) to "manually" count the occurrences of some read. Then, I used the actual
FMIndex::Count method to count it and verified that they were the same. I tested a bunch of random sequences, and every one of them came out correct. So, I'm pretty sure that my rank structure and count methods are correct.
Also did a few benchmarks using the new 2-bit "enhanced" version and repeated some benchmarks from last time:
Test parameters:
- read length = 35
- reference = e coli genome NC_010473, minus two characters (there was one Y and one R...) (length = 4,686,136 bases)
- number of reads = 100,000, generated randomly
- this randomly generated set was copied 1000 times, for a total of 100,000,000 calls to
count.
- random seed = 1234
- times are reported in milliseconds wall time.
- Build version is 64-bit, Release mode (
-O3)
- Memory usage is just the memory used by the rank structure
- There was one $ in the sequence, no N's
| Sampling rate (bases) |
Memory usage (bytes) |
Type |
SSE 4.2? |
Time (ms) |
Ratio to 2-bit |
| 64 |
1,759,632 |
2-bit |
No |
27.071 |
1 |
| 256 |
1,320,312 |
2-bit |
No |
46,726 |
1 |
| 2048 |
1,192,176 |
2-bit |
No |
137,583 |
1 |
| 64 |
1,761,936 |
2-bit "special" |
No |
27,778 |
0.97 |
| 256 |
1,322,640 |
2-bit "special" |
No |
47,574 |
0.98 |
| 2048 |
1,194,728 |
2-bit "special" |
No |
138,833 |
0.99 |
| 64 |
2,931,176 |
3-bit |
No |
31,513 |
0.86 |
| 256 |
2,052,536 |
3-bit |
No |
53,110 |
0.88 |
| 2048 |
1,796,264 |
3-bit |
No |
156,759 |
0.88 |
| 64 |
1,759,632 |
2-bit |
Yes |
19,747 |
1 |
| 256 |
1,320,312 |
2-bit |
Yes |
32,898 |
1 |
| 2048 |
1,192,176 |
2-bit |
Yes |
74,230 |
1 |
| 64 |
1,761,936 |
2-bit "special" |
Yes |
20,471 |
0.96 |
| 256 |
1,322,640 |
2-bit "special" |
Yes |
33,809 |
0.97 |
| 2048 |
1,194,728 |
2-bit "special" |
Yes |
76,269 |
0.99 |
| 64 |
2,931,176 |
3-bit |
Yes |
24,855 |
0.79 |
| 256 |
2,052,536 |
3-bit |
Yes |
37,847 |
0.87 |
| 2048 |
1,796,264 |
3-bit |
Yes |
89,793 |
0.84 |
To do:
- Start doing a
Locate method.
- Maybe do some code cleanup
08/06/10
Yesterday and today, I did a bunch of cleanup on both the rank structure and FM index classes. I pulled some methods out of
StaticDNAStructure::Init so the code can be cleaner. I also rethought the naming scheme of a lot of variables to make the code a little more readable. For the FM index, I also pulled out methods from the
FMIndex::Init method and overloaded the method to take in C++ strings (which are used by
saligner currently). I also added the building of the BWT string directly into the FM index class instead of making it built externally. Currently, only one sequence is supported, but later on, there will be multi-sequence support.
Chris suggeted in an email that we prefetch
ep ranks while doing both counts and locates, which might make it faster. I'll be discussing this with him later.
Finally, I finished the
FMIndex::Locate function today. It works as expected, and I've tested it pretty thoroughly. Locate requires a bunch of LF mappings of each position in the BWT string to travel "backwards" until a position is reached whose position has been indexed. However, I can't actually get the specific character at the given position in the BWT to do the LF mapping. The FM index paper suggests doing an LF on both position
i and
i-1 for each character in the alphabet, since the only one that will differ in LF value is the character I'm currently on. I'm also thinking I could just implement a
select function in my rank structure, which shouldn't be too hard.
To do:
- Benchmarks! I might be able to integrate the index into
saligner without too much trouble, so we can get some pretty accurate comparisons?
- Implement saving/loading of the whole index. Currently, I support saving/loading of the rank structure, and the FM index is just a few more data structures so it shouldn't be too bad.
08/10/10
Implemented a save/load feature for the FM index. I'm also thinking of implementing a "partial" load feature, where only the BWT string is loaded, and the other data structures are still built. The reason for this is that the BWT string is the one that takes the most memory (and maybe time, too) to build and should be constant for all indexes, while the other structures will differ depending on sampling rates and memory restrictions. So, the BWT string can be passed around between machines (with different memory restrictions) easily, while the other data structures can't.
I also did a few preliminary benchmarks, and the times were not great on the
Locate function. I think this might be because we don't implement
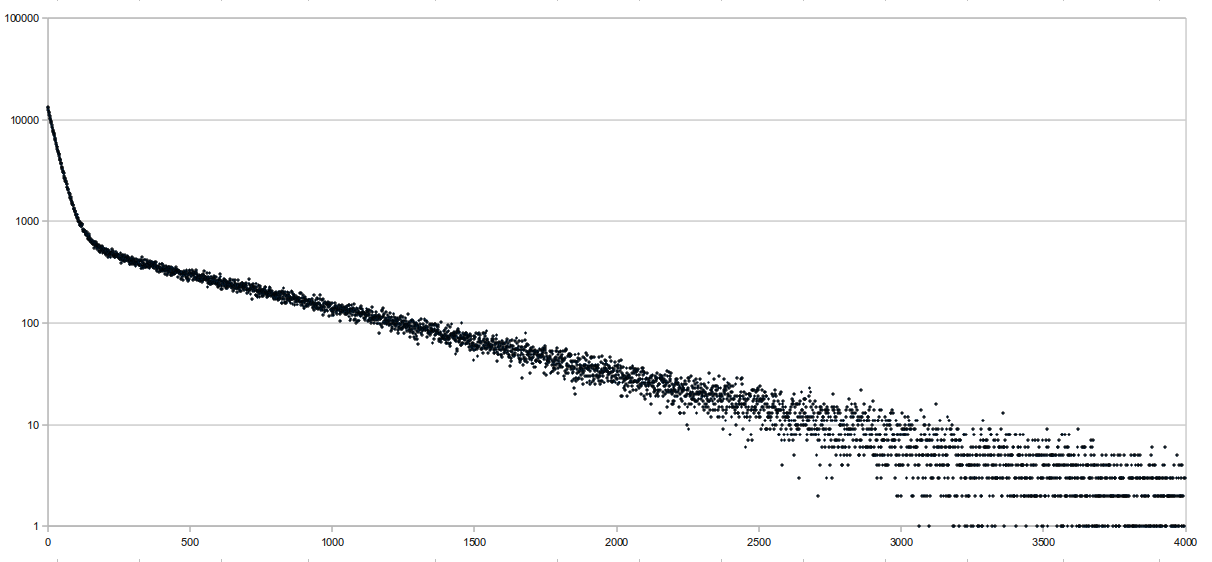
Locate the "proper" way, which guarantees a successful locate within sampling rate number of queries. Following Chris' suggestion, I tried graphing the number of backtracks it takes before a successful location is found on a random readset, and here are the results:
The sequence length was 65,751,813 bases long and consisted of the E. coli genome (4.6 Mbp) repeated multiple times. Reads were randomly generated 10-base sequences, and only the first 10 matches were "located". This was run at a sampling rate of 64 bases, and the average distance to locate was 341.68, which isn't very good. The x axis of the graph represents the distance it takes to locate, and the y axis is the number of aligments with that distance (it's logged). There were a total
- Reference length = 65,751,813 bp (consisting of the E. coli genome repeated multiple times)
- Randomly generated 10-base sequences, only the first 10 alignments were "located"
- Run at a sampling rate of 64 bases for the locate structure
- Average distance = 341.68, which isn't very good
- x-axis is the distance it takes to locate, y-axis is the number of alignments with that distance, semi-logged
- 855,150 locates performed in total
To do:
- Make a "proper" locate structure and compare.
08/11/10
Finished implementing the "proper" locate structure, which guarantees a hit in sampling rate backtracks. I did a few small tests first, where I set the DNA rank structure sampling rate to a constant 64 bits. For aligning 2 million reads a maximum of 1 time, the "proper" version outperforms the previous version by quite a bit, even under roughly the same memory usage (6.9 seconds at sr=64, memory=37MB; vs 25.9 seconds at sr=16, memory=39MB). Even if I up the sampling rate of the "proper" locate to 256 bases (memory usage = 34MB), it performs at comparable speeds.
To do:
- Benchmark more thoroughly
- Test more thoroughly
- Implement a feature to load/save only the BWT string, instead of the whole index, which could change based on different memory usage profiles.
- Get ready for integration?
08/18/10
I just realized I forgot to update this journal in quite a while. So here's a brief overview of what's been happening:
The FM Index is mostly done, but without multiple-reference support. Chris says this should be an easy fix and can be updated later. Running a bunch of tests, the FM index seems quite fast when doing
Locate (roughly 4x faster than bowtie). However, these results aren't the most reliable because there's no post-processing going into it, nor is there quality support. Some interesting points:
- When doing
Locate, the sampling rate of the locate structure (to store positions) matters quite a bit, so this should be as small as possible.
- The
StaticDNASequence sampling rate has some leeway in the beginning (in my tests, from 64-256 bases were okay), but starts affecting the time significantly after that.
- The
StaticBitSequence sampling rate has pretty much no effect, even up to sampling rates of 1024! Unfortunately, changing this sampling rate also affects memory the least.
- Using SSE4.2 (hardware popcount) doesn't have too much of an effect at low
StaticDNASequence sampling rates (expected); at a sampling rate of 64 bases, the SSE-enabled version was about 4% faster, but goes up to 20% higher at a sampling rate of 1024 bases.
Lately, I've been working with Daniel to integrate the new FM index into NGSA, which has taken most of the past two days. Right now, we have the
BowtieMapper working (albeit with a lot of quick hacks, since multi-references isn't done yet), which maps reads with up to 3 mismatches and uses a jump table. Daniel is also going to work on the
BWAMapper, which currently has some problems with unsigned integers and -1's. I'm also going to reorganize the structure of the NGSA library and make a better utilities file.
Finally, I ran some preliminary benchmarks between the new
saligner and
bowtie. The following were run on
NC_010473.fasta, minus the 'R' and the 'Y' character in the genome, using the
e_coli_2622382_1.fq readset. Tests were done on
skorpios in
/var/tmp/, using 64-bit versions of both aligners:
| Aligner |
Match Type |
Flags |
Memory Usage (rough, within a few 1000 B) (B) |
Time (s) |
Notes |
| bowtie |
exact |
-S -v 0 |
6,800,000 |
28.01 |
|
| saligner |
exact |
N/A |
6,500,000 |
18.24 |
locate sr = 16, StaticBitSequence sr = 512, StaticDNASequence sr = 128 |
| saligner |
exact |
N/A |
5,300,000 |
19.07 |
locate sr = 32, StaticBitSequence sr = 512, StaticDNASequence sr = 128 |
Note the times might not match so well because I used
time for bowtie, but the built-in timing function for
saligner, since I haven't gotten the saving/loading integrated yet. Also, I didn't use
valgrind --tool=massif to profile
saligner, because there are some full-text references being kept in memory somewhere, which is really raising the memory usage (I'll have to find and clear those later). The memory reported above is from the FM Index's
FMIndex::MemoryUsage() function, which only reports the theoretical usage, and I kept the memory usage for
saligner on the low side, to account for any extra memory that may be used for other things.
To do:
- Reorganize directory structure
- Fix a bug in the FM Index/=StaticDNASequence=, where Rank0 gives the wrong output in blocks with '$'
- Implement a jump table within FM Index
- Maybe implement multiple sequence support (Chris will get back to me on this)
08/23/2010
Chris gave me a list of things to do for the index as well, and I've been working on those the past few days:
- Have options to make building the locate table, jump table, etc. optional
- Add jumptable support
- Add text extraction support (at least be able to extract from a packed representation)
- Add a
BlockArray class
Currently, I've got the jumptable support done and optional build requirements. I'm also working on some unit tests for the FM index. Finally, the bug from the last post (
Rank0 sometimes fails on blocks with '$') has been fixed.
To do:
- Finish up unit tests
- Add text extraction support from a packed string (to do after block array has been made)
- Add a
BlockArray class
09/01/2010
Had a big discussion with Chris about what the FM index interface should look like and the workflow for building/loading/saving an index, so I thought I'd write it all down before I forget:
-
Init() will now be Build(sampling rates ...), and take in paramaeters for sampling rates, with defaults set. The method will build the index from scratch.
-
Load(file) will be Load(file, sampling rates ...), works the same way as Build, but it loads from a file instead of building from scratch. If sampling rates aren't provided, Load will use the sampling rates specified in the save file; otherwise, it will override the sampling rates, if it can (sampling rates should be a multiple of the ones in the save, and be larger).
-
Save(file) will pretty much do what it's always done, but make sure to save the structure in such a way so that the samples are "skippable"
- I'll have
SetXSamplingRate methods at some point that will allow locate and jump table parameters to be settable. Setting it to 0 will unload everything, while setting it to a different sampling rate will reload the structure. These methods will also have a file that can be passed in (optionally), which allows reloading of the structures just from a file.
- Note that the rank structures (
StaticBitSequence, StaticDNASequence) won't be dynamically changeable, since those require the BWT to rebuild (I could rebuild from a file, but it's a lot of trouble!)
 June 2010 archive
June 2010 archive July 2010 archive
July 2010 archive
 To do:
To do:  To do:
To do: 
