Parameters
Remarks
Example
See Also
Declaration
void __stdcall nbrhdFractal( int xRange, // horizontal range int yRange, // vertical range bool uplink, // are my ancestors my neighbours? bool downlink // are my descendents my neighbours? );
Parameters
- xRange, yRange
- Range of neighbourhood in horizontal and vertical directions, respectively.
- uplink, downlink
- Determines whether links are propagated up the fractal structure to the root or down to the leaves, or both. (Why computer scientists don't make good gardeners: they plant all their trees upside down!)
Remarks
This function is available through the API. It can be called within the model-supplied routine onSimCreate to generate a deterministic, scale-free network. The grid is divided into blocks of xRange×yRange sites linked to their "parents" (the top left site in each block). This pattern is repeated with blocks of xRange2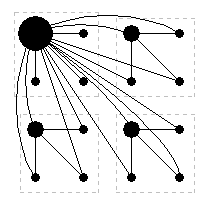
Sample neighbourhood structure for xRange=yRange=2.